PNAS | The Bai Chen team and collaborators revealed the activation mechanism of mGlu2 in the C class GPCR family and important mutation sites through computational simulations.
In May 2024, the Bai Chen team at MoMed Biotech and their collaborators published a research paper online in the Proceedings of the National Academy of Sciences (PNAS) entitled "Exploring the Activation Mechanism of Metabotropic Glutamate Receptor 2". This study not only provides a new perspective for us to deepen our understanding of the potential therapeutic target mGlu2 in the central nervous system diseases but also opens up new avenues for developing more effective treatment strategies.
Background:
Central nervous system diseases, such as schizophrenia and depression, are major health problems globally. The existing treatment methods have limited choices and are accompanied by side effects. Therefore, it is particularly urgent to develop more effective drugs and explore new treatment methods.
The deep understanding of the activation mechanism of mGlu2 is crucial for developing new therapies as it is a potential target for treating these diseases. However, due to the complex molecular activation process of mGlu2, the understanding of its activation mechanism is still limited, which greatly restricts our progress in functional cognition and drug design.
Research Testing:
In this study, the research team used computational biology models to simulate key events in the activation process of mGlu2, including protein conformational changes, agonist binding, Gi protein coupling, and GDP release.
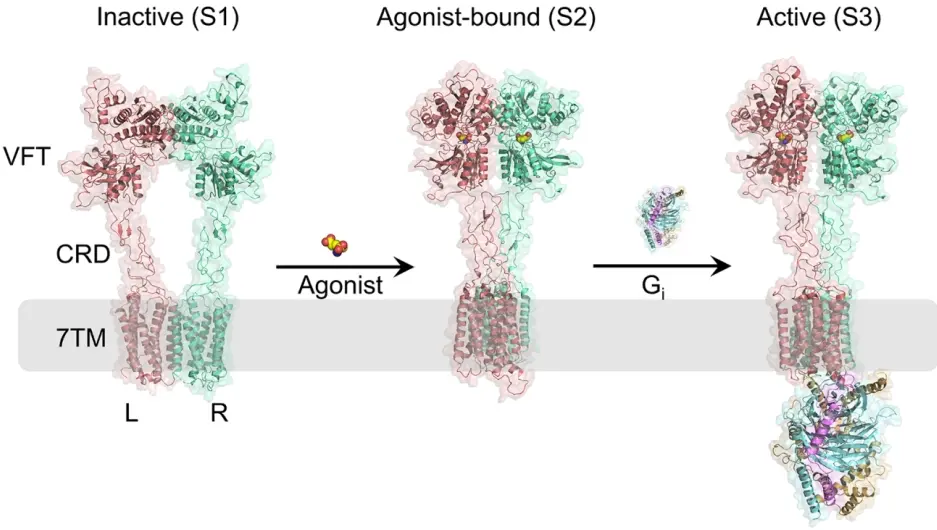
Figure 1. Different conformational states of mGlu2. The L and R subunits of mGlu2 are represented in pink and green, respectively. The agonist is represented using a space-filling model. The Gi protein is represented using three different colors, corresponding to the three subunits of Gi protein: light orange represents the α subunit, blue represents the β subunit, and purple represents the γ subunit.
Through molecular-level simulation and energy analysis, the research team revealed several key energy barriers during the transition of mGlu2 from an inactive state to an active state, and identified that the rate-determining step of the entire activation process occurs during the transition from S2 to S3.
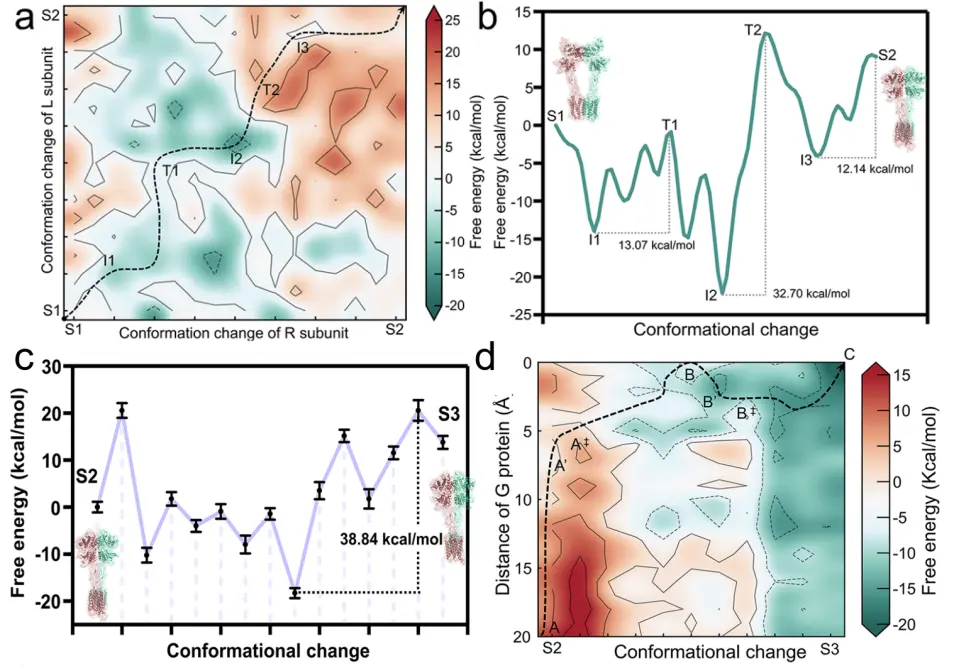
Figure 2. Free energy landscape of conformational changes during mGlu2 activation. (a) Coupled free energy landscape of conformational changes in each subunit of the protein from S1 to S2. The minimum energy path is displayed by a black dashed line. (b) Free energy curve corresponding to the minimum energy path in (a). (c) Free energy curve between S2 and S3 states of mGlu2. (d) Free energy landscape of the binding between G protein and receptor.
In order to study the binding process between the agonist and mGlu2, the research team constructed 195,364 models and plotted the corresponding free energy landscapes. In addition, the research team found that the conformational changes of the two subunits of mGlu2 during the binding process with the agonist are coupled rather than independent processes. The computational results of the research team are consistent with experimental observations, further validating the reliability of the research method.
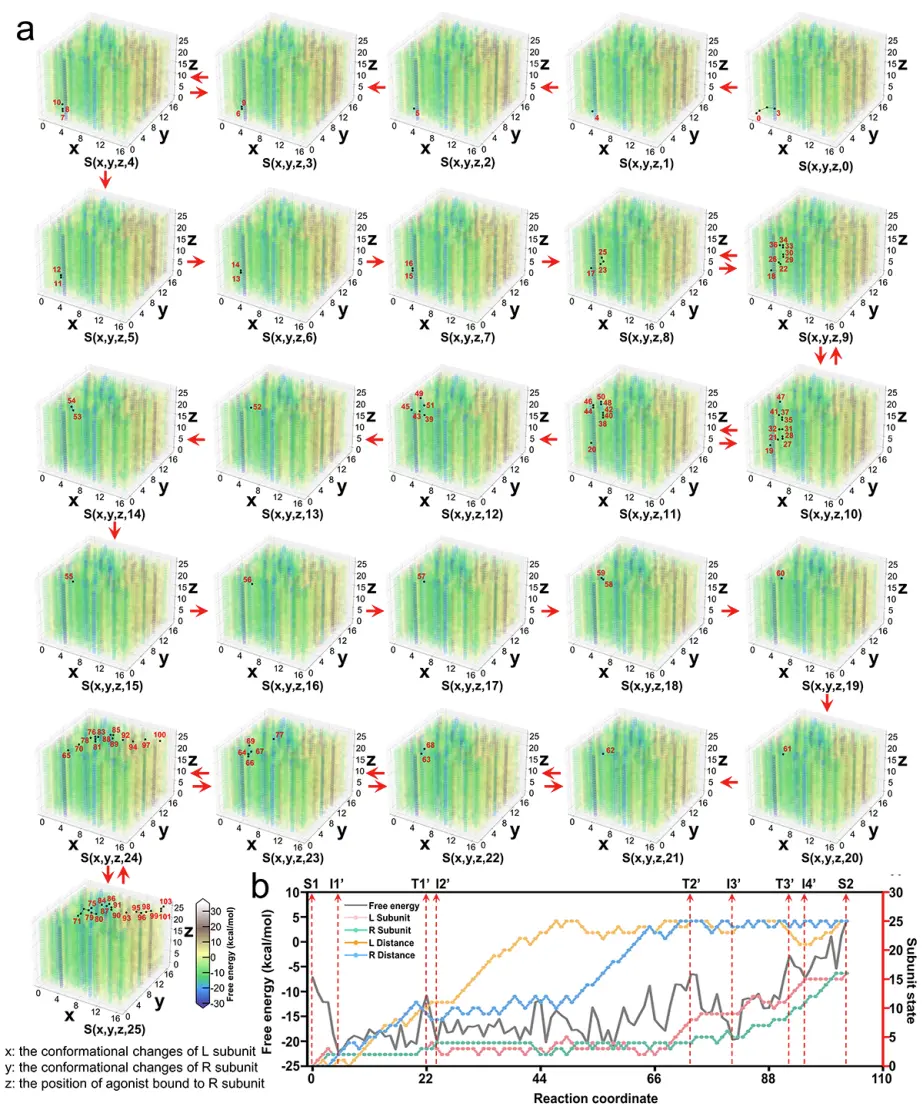
Figure 3. (a) Free energy landscape of conformational changes during the binding of the agonist to mGlu2. The x and y axes represent the conformational changes of the L and R subunits of mGlu2 dimer, and the z axis represents the distance between the pocket in the VFT of the R subunit and the agonist. The distance between the agonist and the pocket in the VFT of the L subunit is fixed and only changes between different cubic boxes. The sampled mGlu2 conformations on the path are marked with black dots and numbered in Arabic numerals in each cubic box. The movement between the red dots is shown as a red arrow inside/outside the box. (b) Optimal conformational pathway of the binding between the agonist and VFT in mGlu2 from S1 to S2 in terms of energy and conformational changes.
In addition, the research team also conducted a mutational effect analysis on the TM6-TM6 interface of the mGlu2 dimer and key residues on the α5 helix of the Gi protein. This helps identify the key residues that influence the activation process of mGlu2 and provides direction for future studies on mutational effects.

Figure 4. Mutational effect analysis of residues in the binding site of mGlu2 and G protein.
In summary, this study not only expands our theoretical understanding of the activation mechanism of Class C GPCRs, but also provides valuable information and inspiration for the design and development of mGlu2 drugs in practical applications. With a deeper understanding of the mGlu2 activation mechanism, we have reason to believe that there will be more innovative therapies for central nervous system diseases in the future, bringing new hope to patients.
The Chinese University of Hong Kong (Shenzhen), University of Science and Technology of China, Zhejiang University, University of Southern California, and MoMed Biotech are the co-collaborating institutions of this paper.
Original paper link: https://doi.org/10.1073/pnas.2401079121
Hot News
-
2022-11-05
The Bai Chen Research Group Publishes Paper in the International Journal of Molecular Sciences
- 2022-12-20
- 2022-12-26
- 2026-02-25




